Deep learning models achieve considerably higher accuracy than traditional computer vision algorithms. When working on image recognition using traditional methods, feature extraction algorithms are tuned by hand which in many cases is a time-consuming procedure. On the contrary, in deep convolutional networks feature engineering is performed automatically – the network learns how to extract the best feature maps on its own and optimizes kernels in subsequent convolutions to preserve only relevant information from images.
So now we don’t need to spend weeks seeking optimal parameters. But it comes with a price. To get adequate results with a complex deep learning model we need large enough datasets. Collecting and annotating big datasets requires a lot of time and financial resources. Moreover, the labeling process itself can be challenging. Synthetic data is a promising alternative to deal with the lack of large enough datasets and to reduce the resources and costs associated with collecting such data [1]. Moreover, it might help institutions to share knowledge, e.g. datasets in highly specialized areas, while protecting individual privacy.
Our goal was to identify microbial colonies on Petri dishes – a typical task in microbiology. The assignment happened to be tough even for trained professionals, because some colonies tend to agglomerate and overlap, thus becoming indistinguishable for non-experts. In this article, we will present an effective strategy to generate an annotated synthetic dataset of microbiological images, that we have already published in Nature Scientific Reports magazine [2]. A generated dataset is then used to train deep learning object detectors in a fully supervised fashion. The generator employs traditional computer vision algorithms together with a neural style transfer method for data augmentation. We show that the method is able to synthesize a dataset of realistic-looking images that can be used to train a neural network model capable of localizing, segmenting, and classifying five different microbial species. Our method requires significantly fewer resources to obtain a useful dataset than collecting and labeling a whole large set of real images with annotations.
We show that starting with only 100 real images, we can generate data to train a detector that achieves comparable results [3] to the same detector but trained on a real, several dozen times bigger microbial dataset [4] containing over 7k images.
Generating a synthetic dataset
Let us now present a detailed description of the method. The goal is to generate synthetic images with microbial colonies that will be later used to train deep learning detection and segmentation models. The pipeline is presented in Fig. 1. Note that the source code with the Python implementation of our generation framework is publicly available.

We start with labeled real images of Petri dishes and perform colony segmentation using traditional computer vision algorithms, including proper filtering, thresholding in CIELab color space, and energy-based segmentation – we use a powerful Chan-Vese algorithm. To get a balanced working dataset, we randomly select 20 images for each of the 5 microbial species (giving 100 images in total) from the higher-resolution subset of the recently introduced AGAR dataset [4].
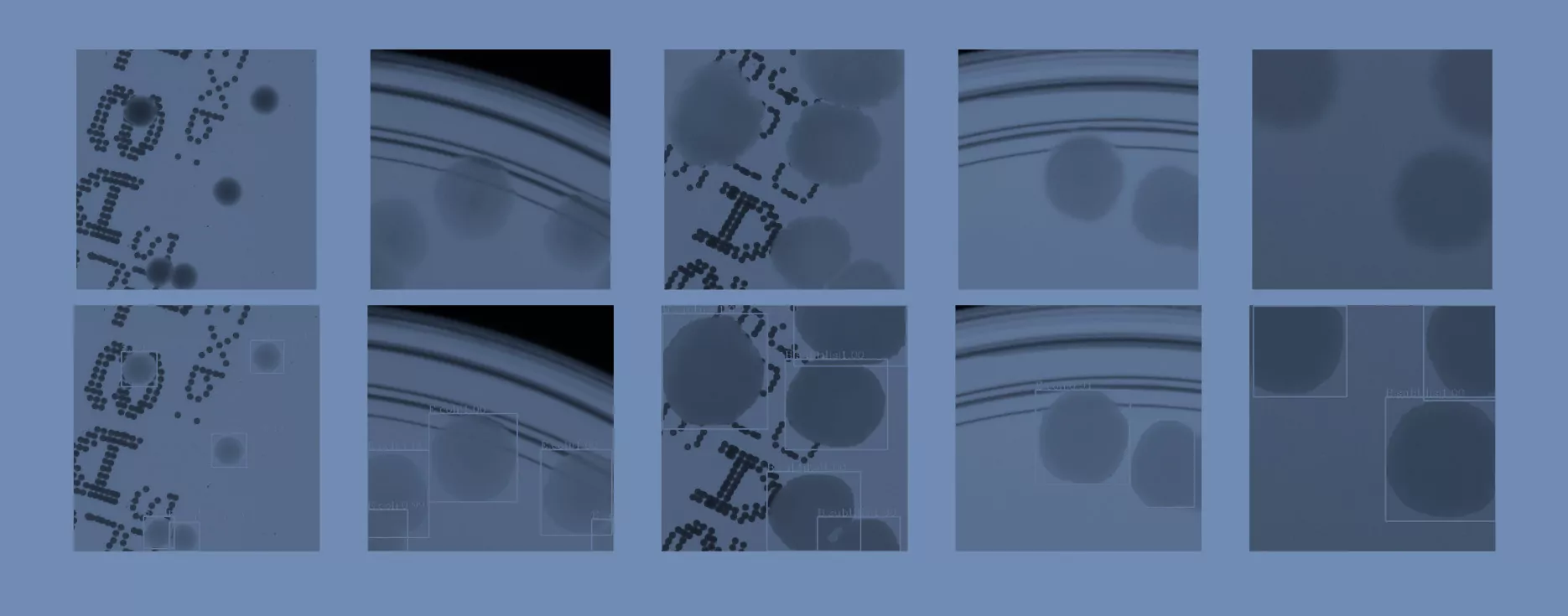
In the second step, the segmented colonies and clusters of colonies are randomly arranged on fragments of an empty Petri dish (we call them patches). We select a random fragment of one out of 10 real empty dish images. We repeat this step a lot of times, placing subsequent clusters in random places, making sure they do not overlap. Simultaneously, we store the position of each colony from the cluster placed on the patch and its segmentation mask, creating a dictionary of annotations for that patch. We present examples of generated synthetic patches in Fig. 2.
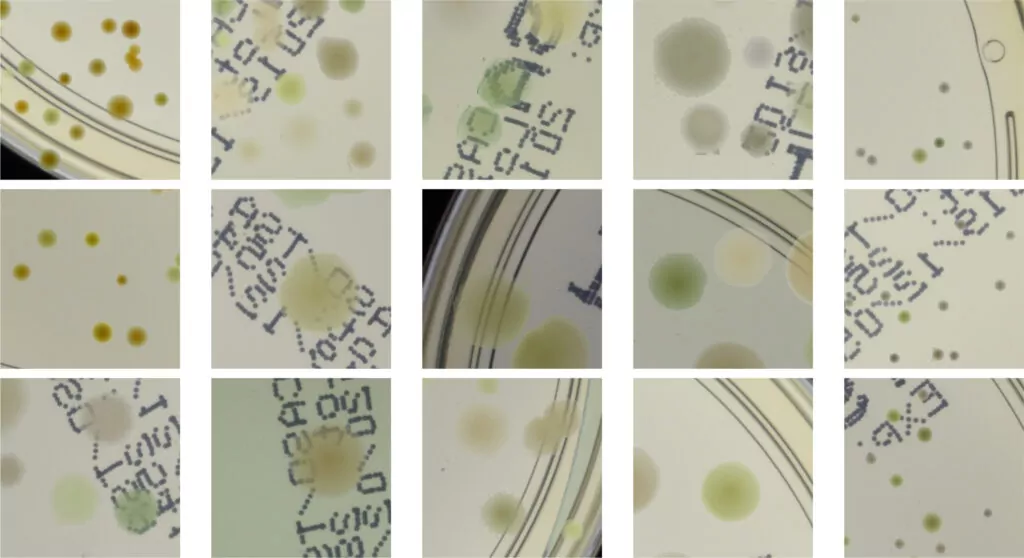
As we can see in Fig. 2, in some situations colonies do not blend well with the background, and their color does not match the background color. To deal with this problem and improve the realism of generated data, in the third step we apply data augmentation using a neural style transfer method. We transfer the style to a given raw patch from one of the selected real images that serve as style carriers. We select 20 real fragments with significantly different lighting conditions to increase the diversity of the generated patches. The exemplary patches after the stylization step are presented in Fig. 3. We use a fast and effective deep learning stylization algorithm introduced in [5]. This method gives us the most realistic stylization of our raw microbial images without introducing any unwanted artifacts.
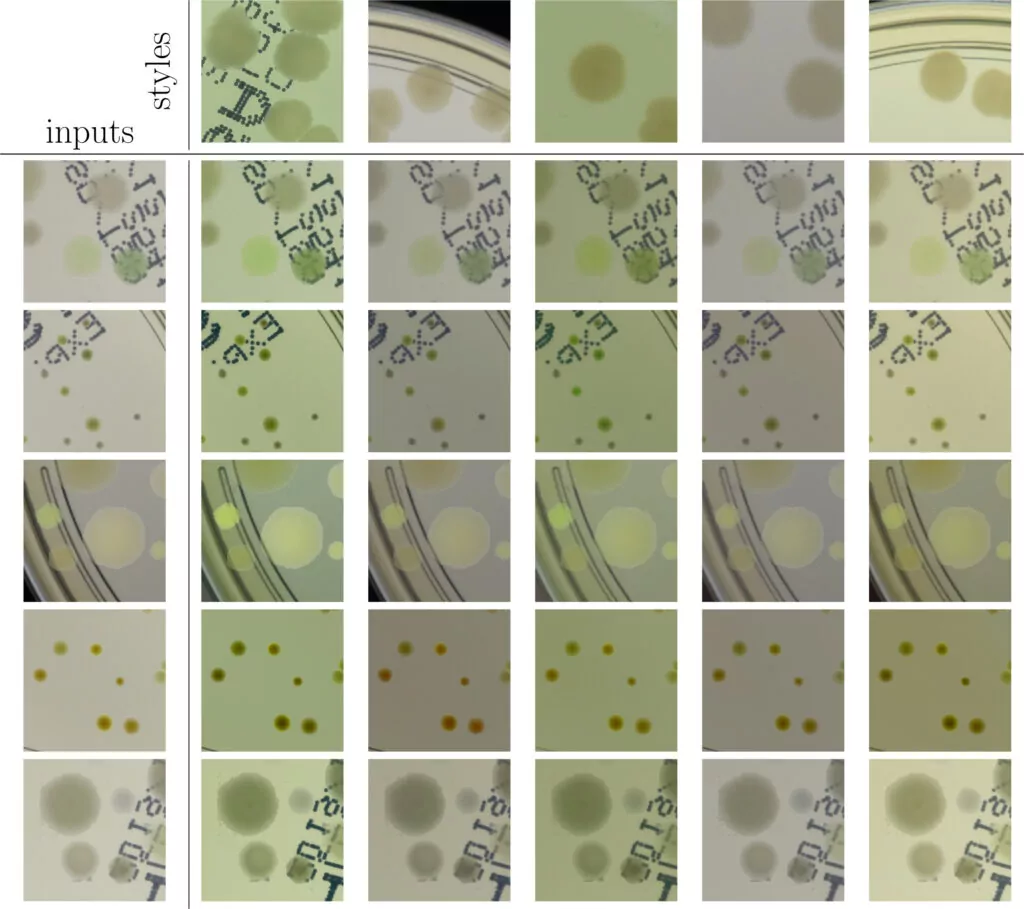

Training deep learning models
Using the method, we generated about 50k patches that were next stylized. The idea behind the conducted experiments was to train a neural network model using synthetic data to detect microbial colonies and then test its performance on real images with bacterial colonies on a Petri dish. We train popular R-CNN detectors using our synthetic dataset. The examples of the Cascade R-CNN [6] detector evaluation on real patches from the AGAR dataset are presented in Fig 4. The model performs quite well detecting microbial colonies of various sizes under different lighting conditions.
Automatic instance segmentation is a task useful in many biomedical applications. During the patch generation, we also store a segmentation mask at a pixel level for each colony. We used this additional information to train a deep learning instance segmentation model – Mask R-CNN [7] which extends the R-CNN detector that we have already trained. The segmentation results for real samples are also presented in Fig. 4. Obtained instance segmentations for different microbial colony types correctly reproduce the colony shapes.
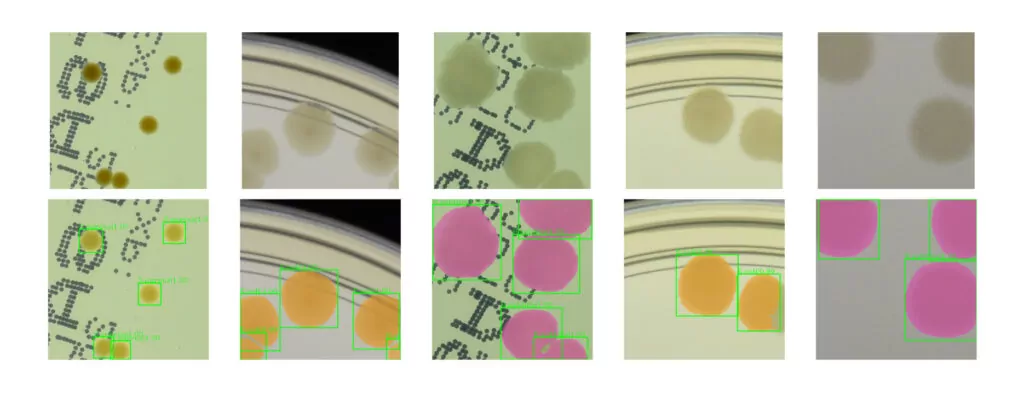
One of the main applications of object detection in microbiology is to automate the process of counting microbial colonies grown on Petri dishes. We verify the proposed method of synthetic dataset generation by comparing it with a standard approach where we collect a big real dataset and train the detector for colony identification and counting tasks.
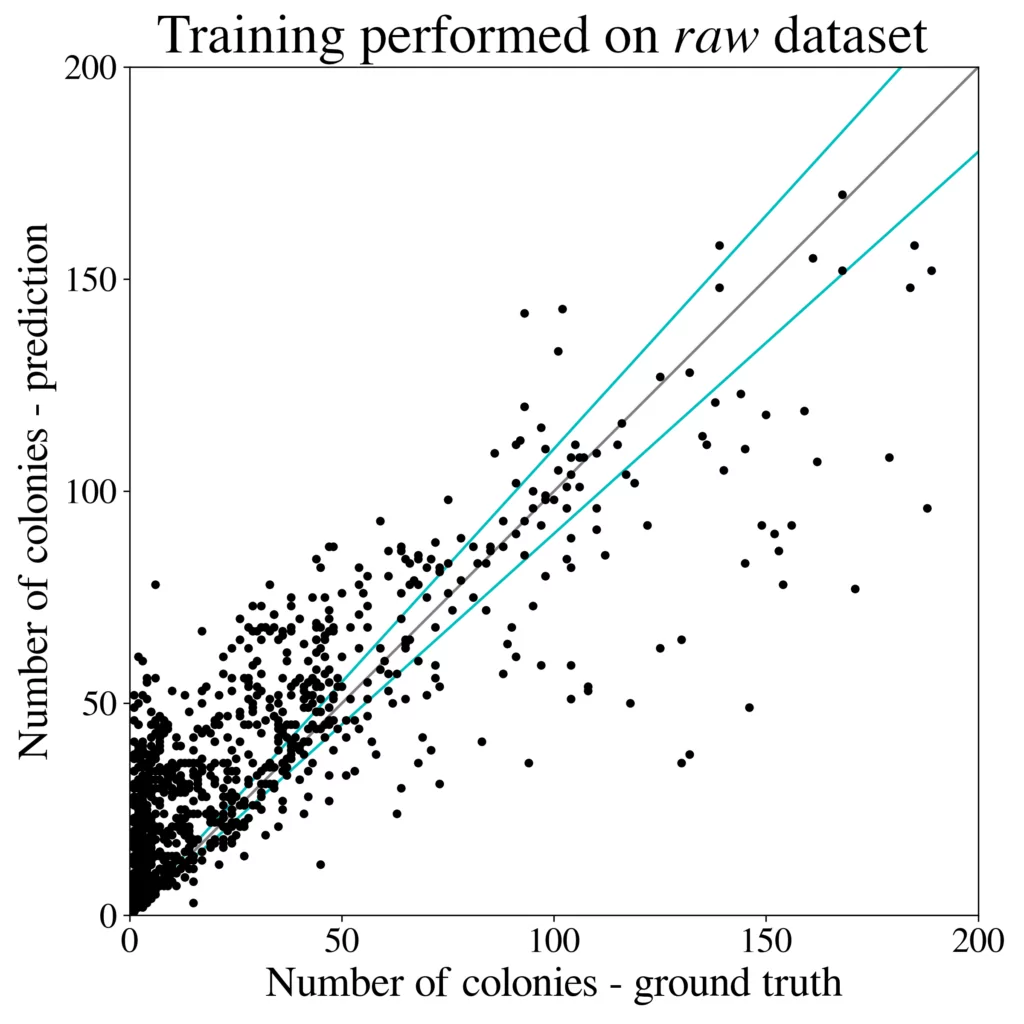
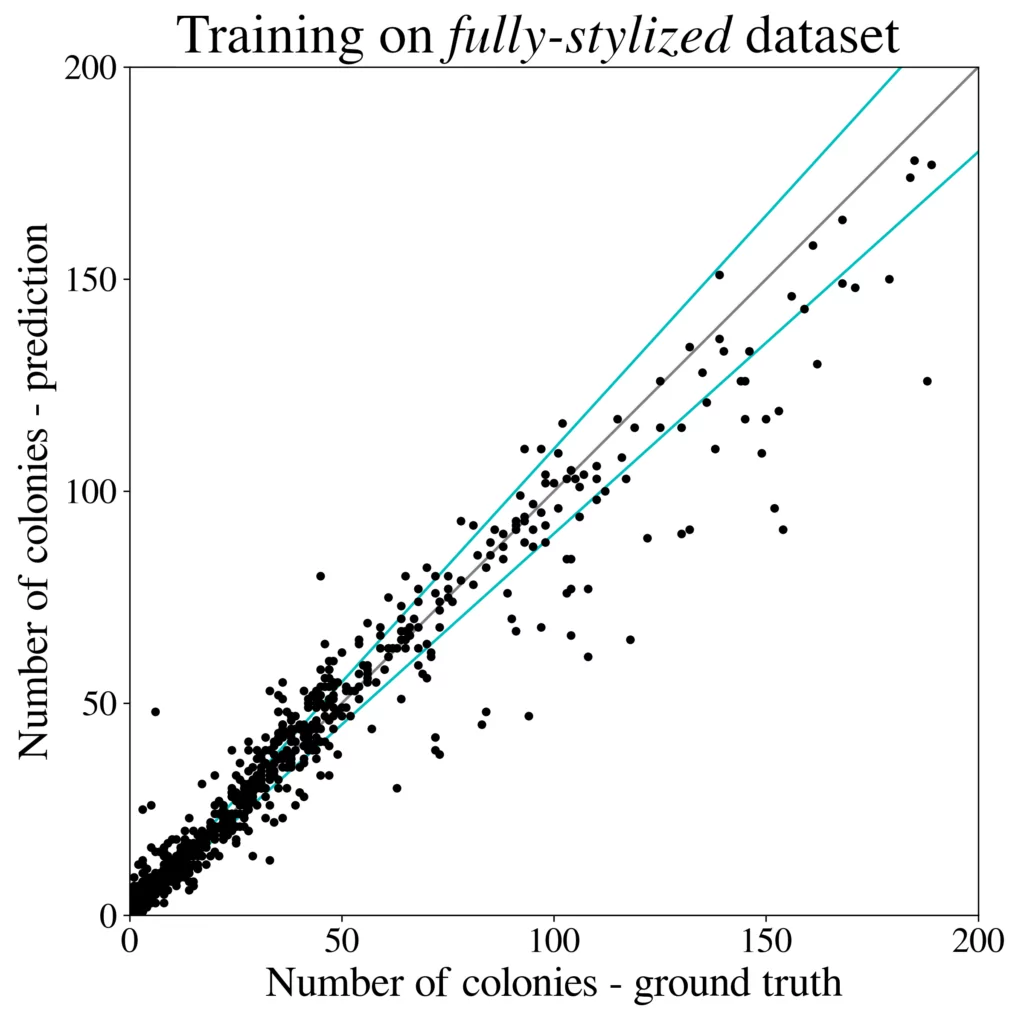
We train the R-CNN detector (Cascade) on a 50k large dataset generated using 100 images from the higher-resolution AGAR subset and test microbial colonies counting in the same task as performed in [4]. Results are presented in Fig. 5 (right). It turns out that the detection precision and counting errors for the synthetic dataset are only slightly worse [3] than for the same detector but trained on the whole big dataset containing over 7k real images giving about 65k patches. It is also clear that introducing style transfer augmentation improves the detection quality greatly, and without the stylization step the results are rather poor – see results for the raw dataset, i.e. obtained without the stylization step, in Fig. 5 (left).
Summary
We introduced an effective strategy to generate an annotated synthetic dataset of microbiological images of Petri dishes that can be used to train deep learning models in a fully supervised fashion. By using traditional computer vision techniques complemented by a deep neural style transfer algorithm, we were able to build a microbial data generator supplied with only 100 real images. It requires much less effort and resources than collecting and labeling a large dataset containing thousands of real images.
We prove the usefulness of the method in microbe detection and segmentation, but we expect that being flexible and universal, it can also be applied in other domains of science and industry to detect various objects.
References
[1] https://blogs.nvidia.com/blog/2021/06/08/what-is-synthetic-data
[2] J. Pawłowski, S. Majchrowska, and T. Golan, Generation of microbial colonies dataset with deep learning style transfer, Scientific Reports 12, 5212 (2022).
https://doi.org/10.1038/s41598-022-09264-z
[3] Detection mAP = 0.416 (bigger is better), and counting MAE = 4.49 (smaller is better) metrics, compared with mAP = 0.520 and MAE = 4.31 obtained the same detector but trained using the AGAR dataset [4].
[4] S. Majchrowska, J. Pawłowski, G. Guła, T. Bonus, A. Hanas, A. Loch, A. Pawlak, J. Roszkowiak, T. Golan, and Z. Drulis-Kawa, AGAR a Microbial Colony Dataset for Deep Learning Detection (2021). Preprint available at arXiv [arXiv:2108.01234].
[5] Ming Li, Chunyang Ye, and Wei Li, High-resolution network for photorealistic style transfer (2019). Preprint available at [arXiv:1904.11617].
[6] Z. Cai, and N. Vasconcelos, Cascade R-CNN: Delving into high quality object detection, IEEE/CVF Conference on Computer Vision and Pattern Recognition, 6154–6162 (2018).
[7] K. He, G. Gkioxari, P. Dollár, and R. Girshick, Mask R-CNN, IEEE International Conference on Computer Vision (ICCV), 2980–2988 (2017).
Project co-financed from European Union funds under the European Regional Development Funds as part of the Smart Growth Operational Programme.
Project implemented as part of the National Centre for Research and Development: Fast Track.